
Welcome to pycoMeth documentation
DNA methylation analysis downstream to Nanopolish for Oxford Nanopore DNA sequencing datasets
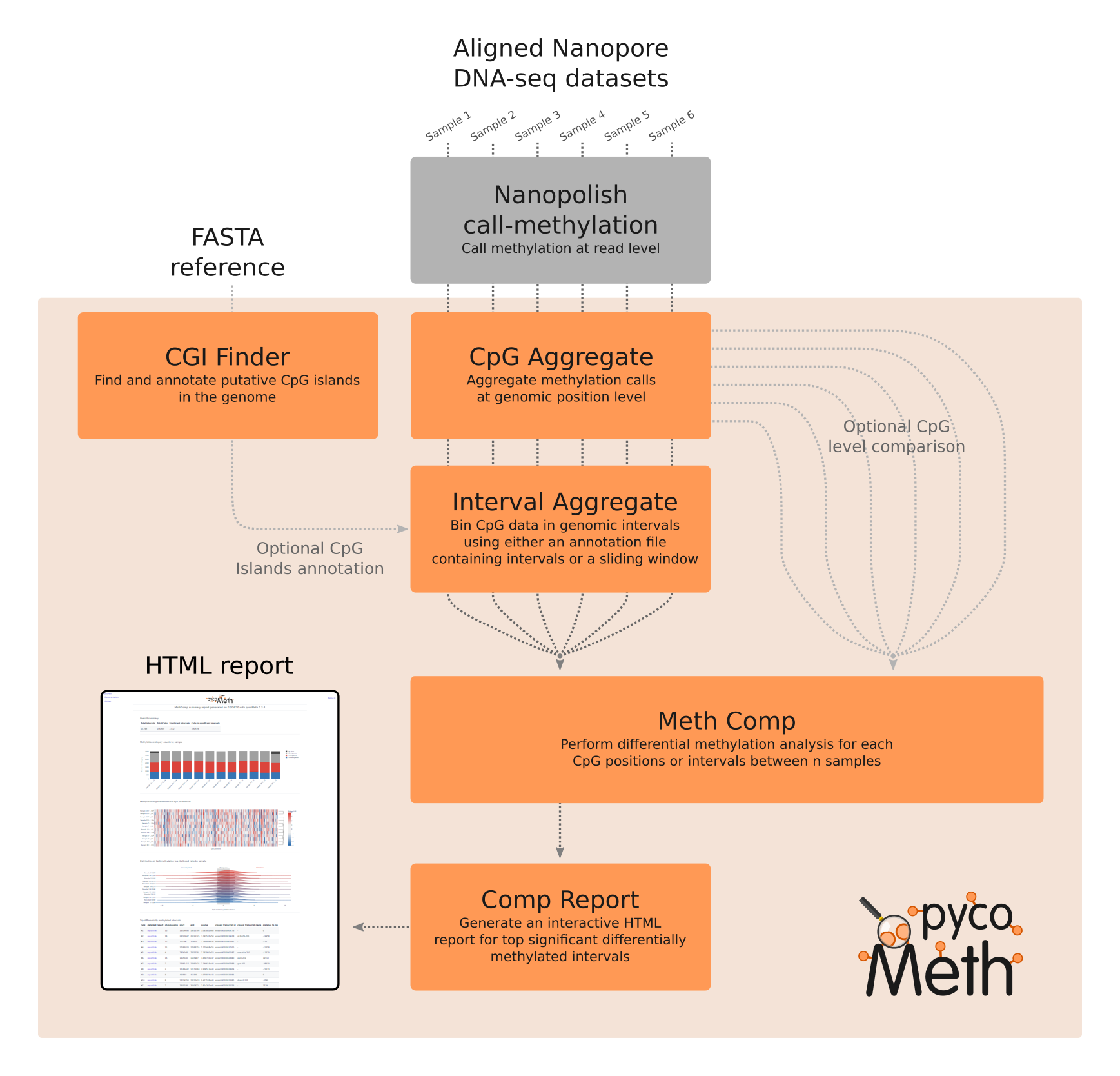
pycoMeth can be used for further analyses starting from the output files generated by Nanopolish call-methylation. The package contains a suite of tools to find CpG islands calculate the methylation probability at CpG dinucleotide or CpG island resolution across the entire genome and to perform a differential methylation analysis across multiple samples. Finally an interactive HTML report can be generated to visualize significant intervals.
The package contains the following modules:
- CGI_Finder: Find CpG islands in a fasta file by using a sliding window and merging overlapping windows satisfying the CpG island definition.
- CpG_Aggregate: Aggregate the output of
nanopolish call-methylationat genomic position level. - Interval_Aggregate: Bin the output of
pycoMeth CpG_Aggregatein genomic intervals, using either an annotation file or a sliding window. - Meth_Comp: Compare Methylation level between several samples from either
pycoMeth CpG_AggregateorpycoMeth Interval_Aggregate. - Comp_Report: Generate an interactive HTML report for top significant differentially methylated intervals found with
pycoMeth Meth_Comp
pycoMeth generates extensive tabulated reports and BED files which can be loaded in a genome browser. In addition, an interactive HTML report of differentially
methylated intervals/islands can also generated at the end of the analysis.
Methplotlib developed by Wouter de coster is an excellent complementary tool to visualise and explore methylation status for specific loci.
Please be aware that pycoMeth is a research package that is still under development.
The API, command line interface, and implementation might change without retro-compatibility.
pycoMeth workflow
pycoMeth example output IGV rendering

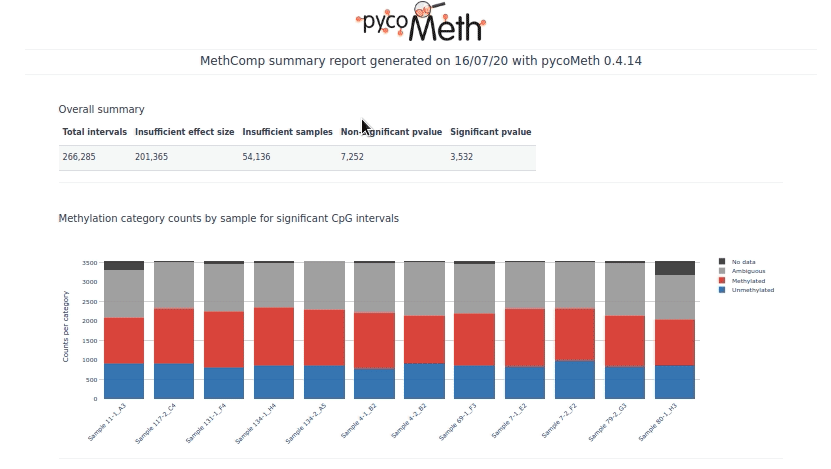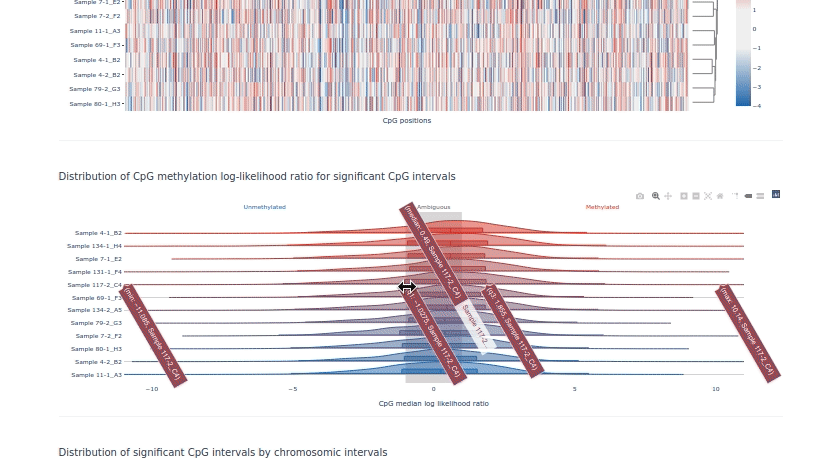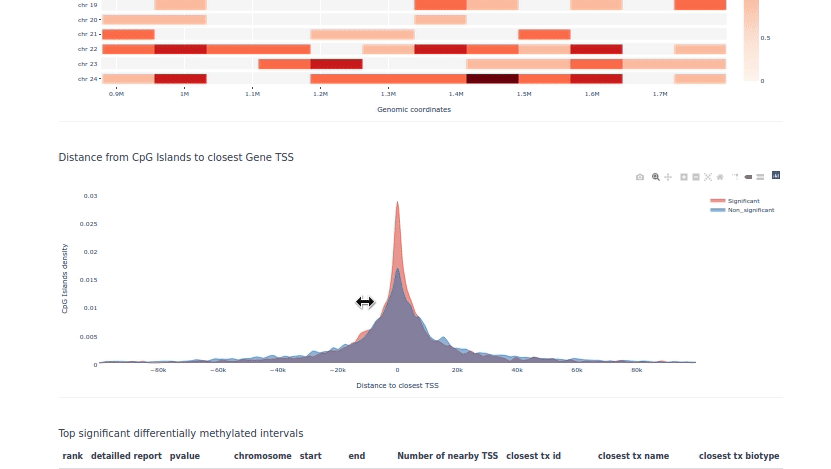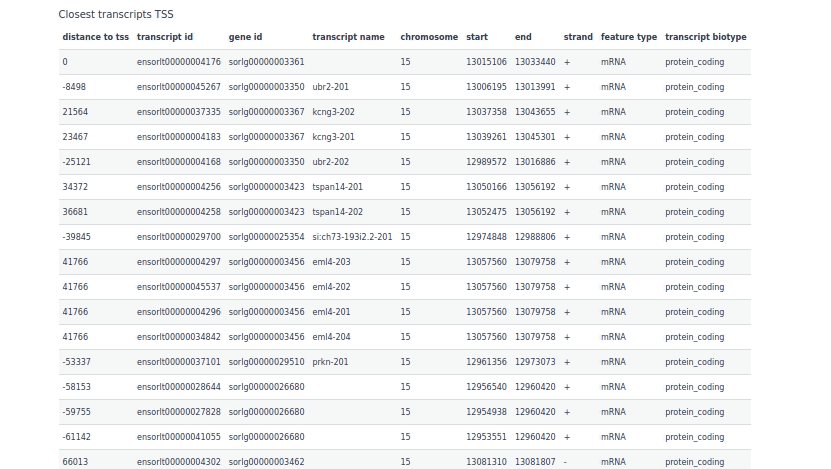
pycoMeth example HTML report
Citing
The repository is archived at Zenodo. If you use pycoMeth please cite as follow:
Adrien Leger. (2020, January 28). a-slide/pycoMeth. Zenodo. https://doi.org/10.5281/zenodo.3629254
Authors
- Adrien Leger - aleg {at} ebi.ac.uk

